I have a landuse shapefile (contains data and script) that plots differently in Base and ggplot, the reason for which remains a myster after countless hours of playing with it.
I read in both shapefiles as follows, since catchment_landuse does not have an ID column, I add one:
if (!require("pacman")) install.packages("pacman")
pacman::p_load(tidyverse, raster, broom, rstudioapi)
#setwd to file location
setwd(substr(rstudioapi::getActiveDocumentContext()$path, 1, rev(gregexpr("/", rstudioapi::getActiveDocumentContext()$path)[[1]])[1]))
catchment_industry <- readOGR('.', 'catchment_industry')
catchment_landuse <- readOGR('.', 'catchment_landuse')
catchment_landuse@data$id <- 1:nrow(catchment_landuse@data)
Is there actually a difference in
catchment_landuse@data$id <- 1:nrow(catchment_landuse@data)
and
catchment_landuse$id <- 1:length(catchment_landuse$id)
?
My code for plotting the thing in base:
base_plot_spdf <- function(spdf){
# get classes for for-loop and sort alphabetically
classes <- sort(unique(spdf$Class))
# rainbow color vector
class_colors = rainbow(length(classes))
spdf_bbox <- bbox(spdf)
# enable legend outside of plot
par(mar=c(5.1, 4.1, 4.1, 8.1), xpd=TRUE)
#expanded empty plot for legend
plot(NULL, xlim = spdf_bbox[1, ], ylim = spdf_bbox[2, ], ylab = "", xlab = "")
# plot polygons that belong to specific classes
for(i in 1:(length(classes))){
plot(
subset(spdf, Class == classes[i]),
col = class_colors[i],
add = TRUE
)
}
# legend
legend("topright", inset=c(-0.25,0), legend=classes, col = class_colors, lwd=c(2.5,2.5) )
}
base_plot_spdf(catchment_landuse)
gives

to make this plottable in ggplot and keep the attributes, I do
spdf_to_df <- function(spdf){
tidy(spdf) %>%
merge(spdf, by = 'id') %>%
as.tibble
}
catchment_landuse_df <- spdf_to_df(catchment_landuse)
Then I use this code for actual plotting:
plot_spatial_df <- function(df){
ggplot(df) +
geom_polygon(aes(
x = long,
y = lat,
group = group,
col = Class,
fill = Class)
) +
scale_fill_manual(values = rainbow(length(unique(df$Class)))) +
theme_minimal() +
coord_equal()
}
plot_spatial_df(catchment_landuse_df)
Which gives
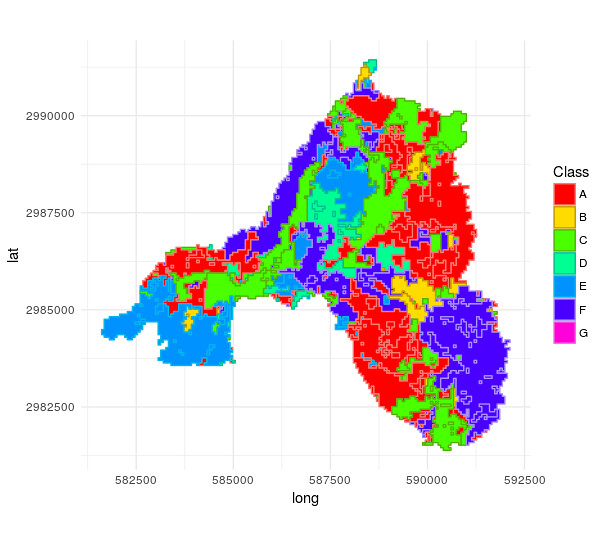
as you can see, especially in the lower left and lower right classes do differ completely. What is the reason for this?
I do not need the "id" column that I create for base graphics, however for ggplot I need to do the "self-join" on the "id" column to keep the attributes.
Also, why does one id get lost somewhere?
length(unique(catchment_landuse_df$id))
[1] 575
How does the @data of the spdf know to which polygons a row in @data refer? What is the link for that?
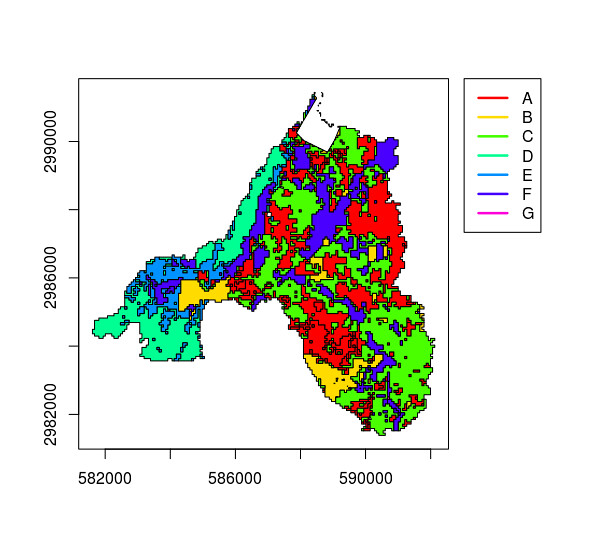
Now I want to substract the second shapefile, catchment_industry from that, keeping all attributes in catchment_landuse:
catchment_landuse_without_industry <- erase(catchment_landuse, catchment_industry)
Base produces the output I expect:
However, ggplot now does really weird things:
catchment_landuse_without_industry_df <- spdf_to_df(catchment_landuse_without_industry)
plot_spatial_df(catchment_landuse_without_industry_df)
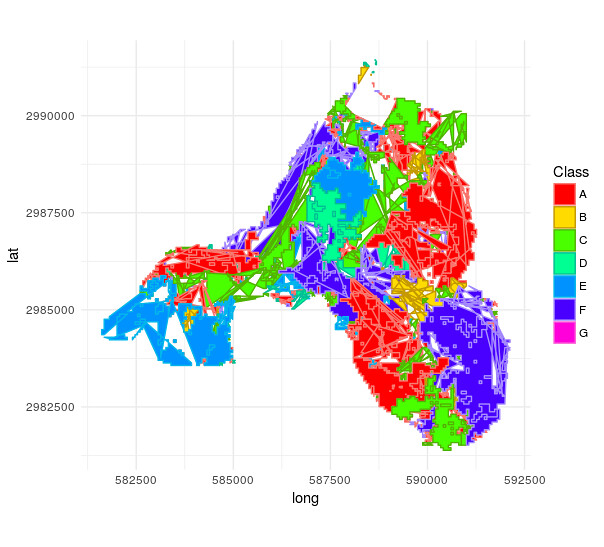
Is the underlying data corrupted in any way or does the mistake solely stem from wrong plotting?
Best Answer
I got a small step closer to the solution of the first problem. The
tidyfunction needsregion = 'id'as argument:This then results in:
However, note that in the larger patches on the lower right and lower left especially there should be small other polygons drawn, which seem to be swallowed somehow.
Also, adding the id column in the following preserves the number of ids after converting the SPDF to a DF